Why Subscribe?✅ Curated by Tommy Tang, a Director of Bioinformatics with 100K+ followers across LinkedIn, X, and YouTube✅ No fluff—just deep insights and working code examples✅ Trusted by grad students, postdocs, and biotech professionals✅ 100% free
R, Python, or Unix? The Bioinformatics Toolkit You Need
|
Hello Bioinformatics lovers, Whether you're just starting or deep into bioinformatics, one question always looms large: R vs Python: Which Should You Learn?Many beginners ask me this, and honestly, the answer depends on your needs. Let me share my journey: I started learning Unix commands over a decade ago, followed by Python through Python for Absolute Beginners. While Python taught me programming fundamentals (like Why? At the time, Python lacked user-friendly libraries for working with data tables (pandas never ticked for me). My work required handling large spreadsheets, so I turned to R. R's built-in support for dataframes made it perfect for bioinformatics tasks. And with the tidyverse, complex data wrangling became simple. Even today, ggplot2 remains one of the best tools for visualizations in bioinformatics. R’s Bioconductor ecosystem is another game-changer. From RNA-seq (think DESeq2) to single-cell analysis, it offers countless well-tested packages tailored for biologists. But R has limitations:
On the other hand, Python is a general-purpose language. While it has fewer specialized bioinformatics tools, it excels in deep learning, algorithm development, and scalability. So, R or Python?Here’s my advice:
Ultimately, both languages are valuable. Learn one well, and the other will come more easily! Why Every Bioinformatician Needs Unix SkillsWhile R and Python are great for scripting and analysis, bioinformatics revolves around text files. From FASTA to SAM files, everything is text-based, and Unix commands are essential for wrangling them efficiently. Here are a few Unix tricks every bioinformatician should know: 1️⃣ Reverse complement a sequence in one line:
Output:
2️⃣ Convert a comma-delimited file to tab-delimited:
Why? Many bioinformatics tools expect tab-delimited input, like BED files. A simple Unix command can reformat your data without the need for Excel or fancy GUIs. 3️⃣ Split a column like
Unix commands like Bottom LineWhether you're wrangling data with Unix, analyzing it in R, or scaling up with Python: Tip: Want to learn more Unix tricks? I’ll be sharing one every week—stay tuned! Happy learning, Tommy aka. Crazyhottommy Other posts from the past week:
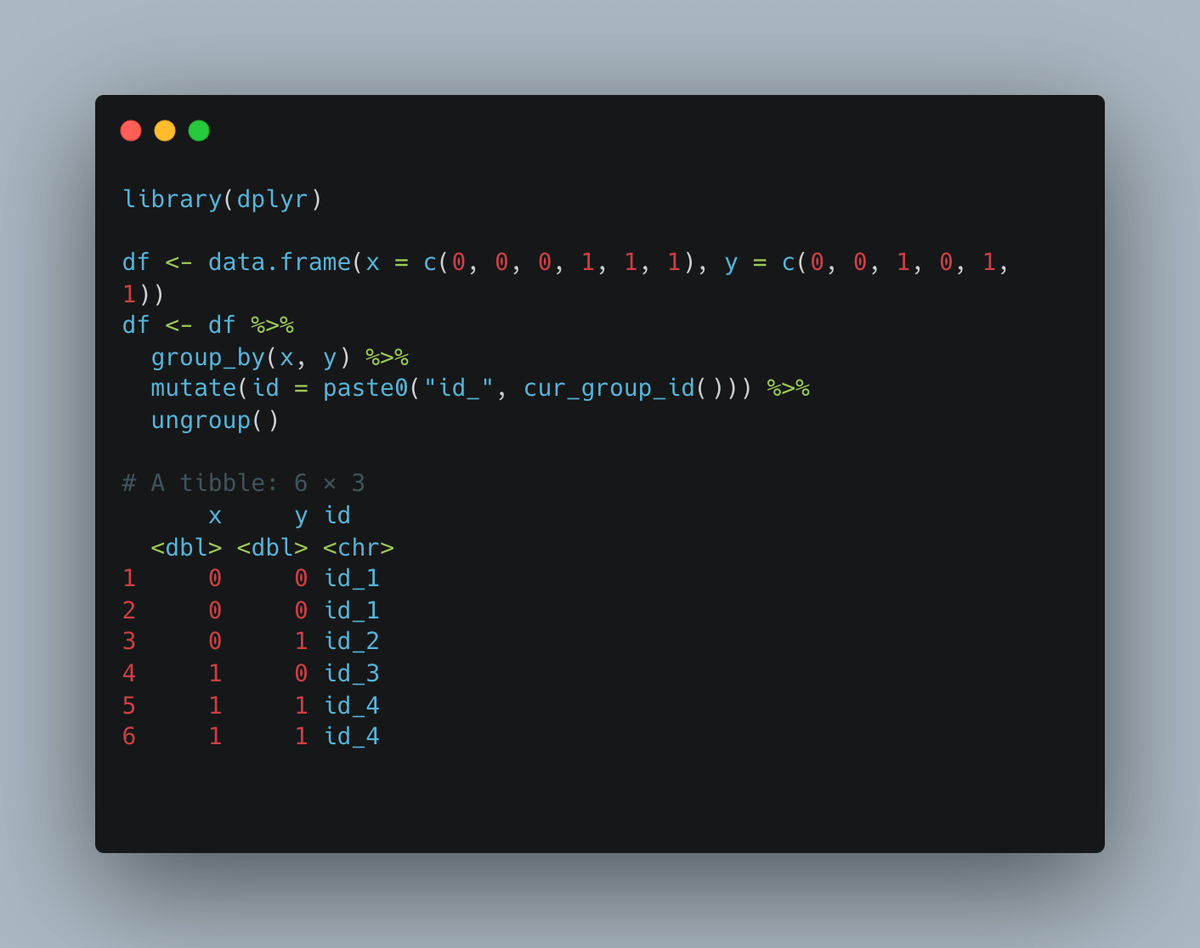
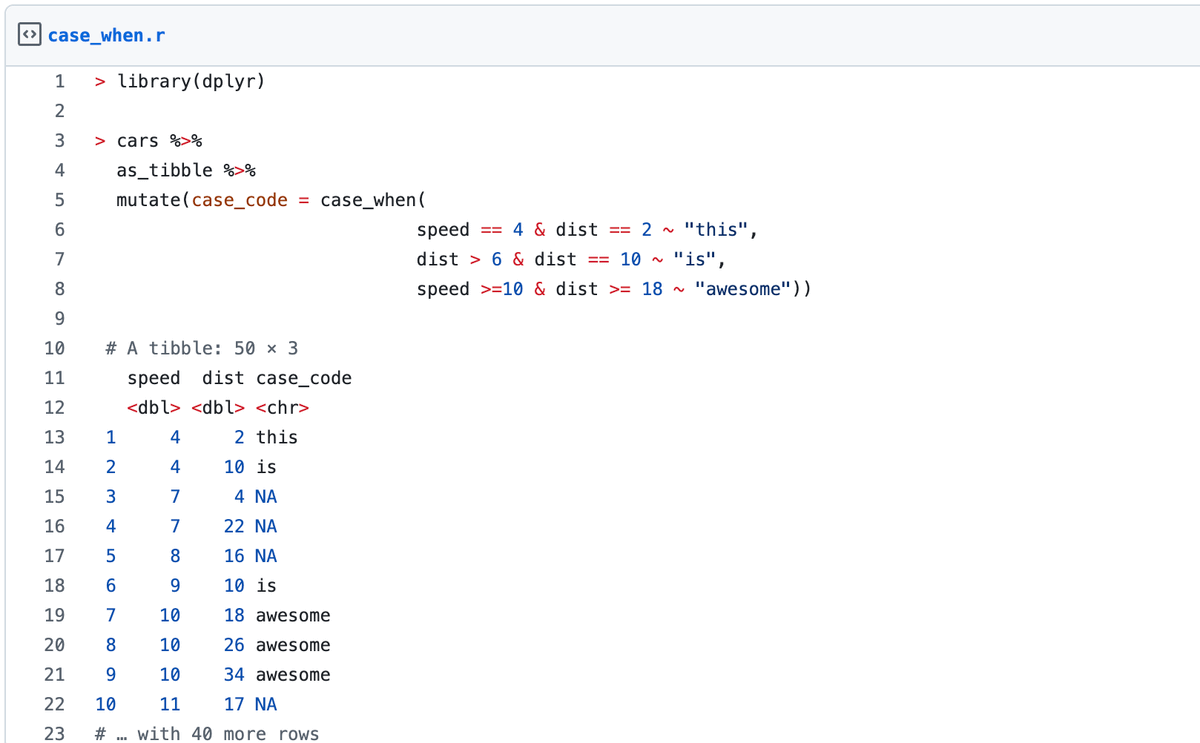
2. Rstats trick: use case_when(). I use it almost every week. No more nested if else
3. A popular course on Bash scripting on Linux. 4. When should you use a package vs. solving a problem from scratch in bioinformatics? I get asked this a lot. Here’s my approach. 5. What happens when biology and computer science collide? A conversation with a colleague revealed just how different these fields can be. Here's why understanding both is crucial for the future of research. PS: I made this youtube video on how to get the sum of exon length for each gene using Bioconductor annotation packages. PPS: If you want to learn Bioinformatics, there are other ways that I can help:
Stay awesome! |
Chatomics! — The Bioinformatics Newsletter
Why Subscribe?✅ Curated by Tommy Tang, a Director of Bioinformatics with 100K+ followers across LinkedIn, X, and YouTube✅ No fluff—just deep insights and working code examples✅ Trusted by grad students, postdocs, and biotech professionals✅ 100% free