Why Subscribe?✅ Curated by Tommy Tang, a Director of Bioinformatics with 100K+ followers across LinkedIn, X, and YouTube✅ No fluff—just deep insights and working code examples✅ Trusted by grad students, postdocs, and biotech professionals✅ 100% free
Seurat and Scanpy log2Fold change discrepancy for single cell data
|
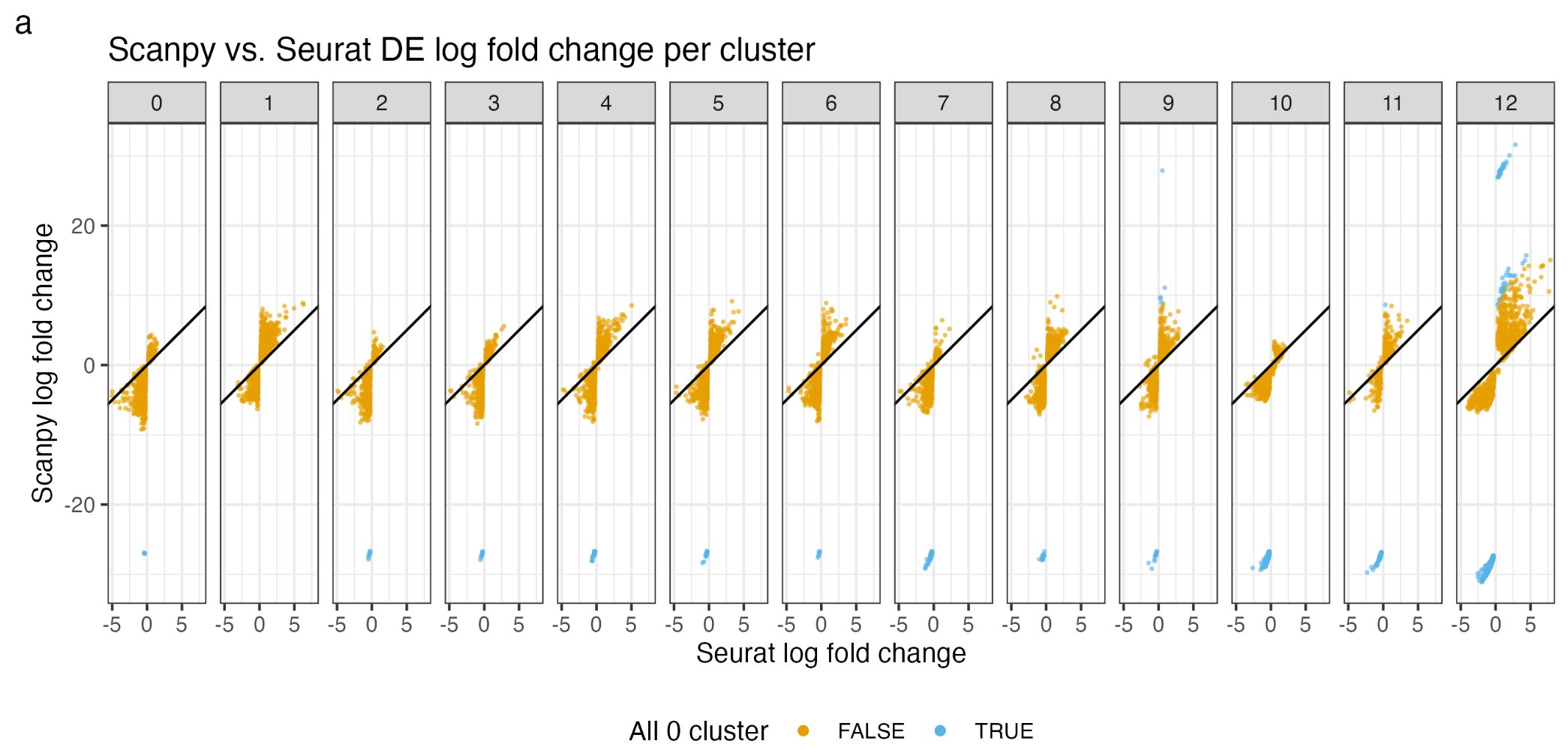
Hello Bioinformatics lovers, If you work on single-cell RNAseq data, do you know Seurat (in R) and scanpy (in python) had a discrepancy in calculating log2Fold change for marker genes per cluster?
Read more details in this blog post: https://divingintogeneticsandgenomics.com/post/do-you-really-understand-log2fold-change-in-single-cell-rnaseq-data/ In this video, I show you how to use google gemini multi-modal to write math formula in Latex by feeding a screenshot to it: Other useful resources this week:
Happy Learning! Tommy Let's connect on twitter and Linkedin! |
Chatomics! — The Bioinformatics Newsletter
Why Subscribe?✅ Curated by Tommy Tang, a Director of Bioinformatics with 100K+ followers across LinkedIn, X, and YouTube✅ No fluff—just deep insights and working code examples✅ Trusted by grad students, postdocs, and biotech professionals✅ 100% free